Description
SWGIS v2.0 is the modified version of the SeqWord Genomic Island Sniffer. This version is specifically optimized for predicting genomic islands in eukaryotic genomes. SWGIS v2.0 was tested on several eukaryotic species of different lineages. All identified genomic islands were deposited in the EuGI database.
Download SWGIS v2.0
Installation
The program needs no installation. Download swgisv2.0. The file contains a Python version of the program compatible with all linux operating systems with Python 2.5 installed. Unzip the file to a selected directory. A folder swgisv2.0 will appear with several files inside and two subordinate folders input and output. To process genomic DNA sequences in FASTA or GenBank formats copy them to the folder input and run SeqWordSniffer.py. The following will appear in the command line:
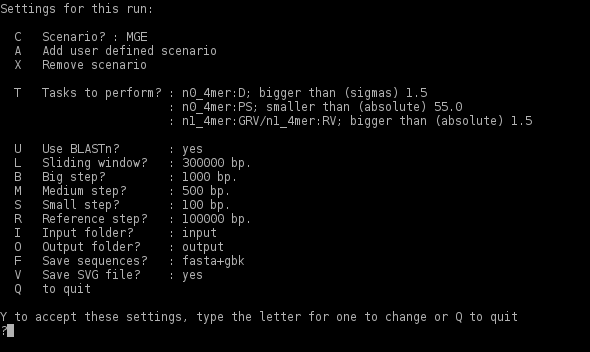
Running swgisv2.0
To change parameters toggle the different letters. to accept defaults press <'Y'> and <'Enter'>. the following will appear in the command line (as an example):
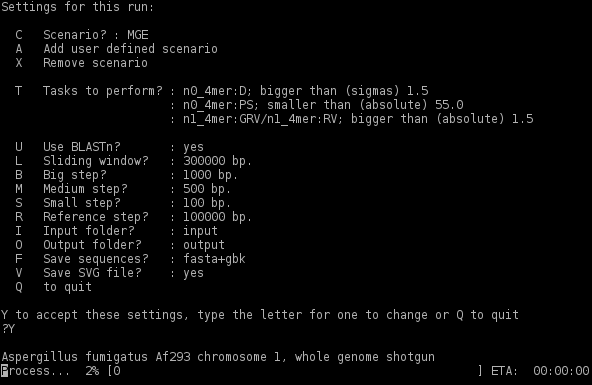
input and output files
The program sequentially process all files of genomic DNA sequences in FASTA ('.FNA','.FAS','.FST','.FASTA') or GenBank ('.GBK','.GB') formats from the input directory and saves the results as in the output directory. Several types of output files may be saved:
- text output file (extension .OUT)
- fasta file of the selected genomic fragments (extension .FAS), toggle by <'F'>
- genbank file for each selected genomic fragments with the annotation data (extension .GBK), toggle by <'F'> but available only when a source GenBank file is processed
- Graphical file with selected fragments mapped over the linear chromosome (extension .SVG), toggle by <'V'>
Options to improve prediction
There is the option to us a blastn algorithm against a rrna database to check if the selected genomic fragments contain rrn clusters. use the <'u'> option to toggle
Edition of the task list
The user may change the default options. Change the set of the OU statistical parameters the program calculates to identify the genomic fragments with <'T'> toggle
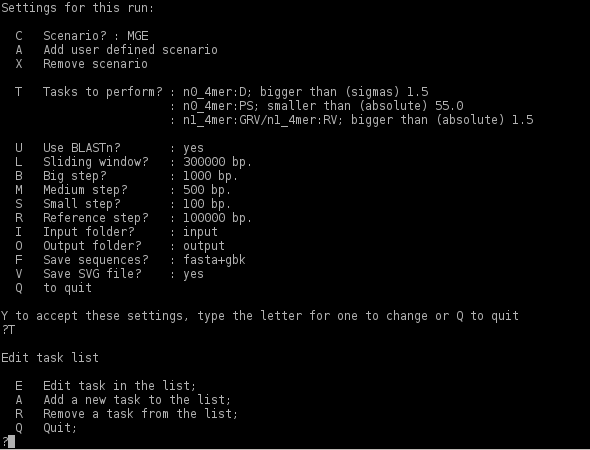
Each task is presented by a line defining the task category and the condition used to select the genomic fragments. Remember that the fragment will be selected only if it meets all set conditions. To remove a condition toggle <'R'>, then select the number of the task to remove it from the list. To return to the main menu toggle <'Q'>.
Setting task conditions
To edit the condition of one of the tasks toggle <'E'>. Then type the number of the task to edit and press <'Enter'>. A submenu of edit options will appear as shown below:
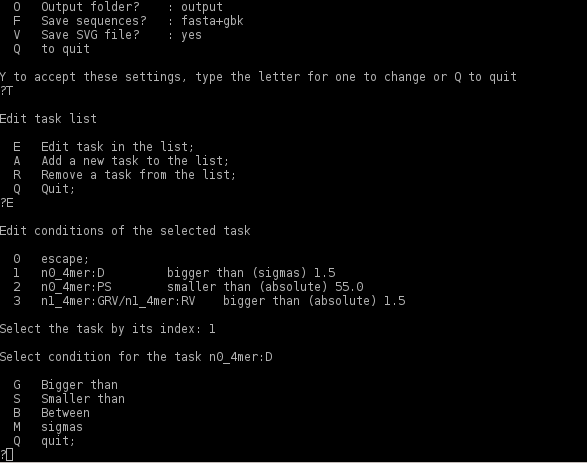
toggle <'M'> to choose the type of the threshold values:
- sigmas - to set the threshold values in sigmas of the normal distribution;
- fraction - to set the threshold as a fraction of the total number of genomic fragments;
- absolute - to use as the threshold an absolute value of the OU statistical paramenetrs.
To choose the type of comparison,- bigger than, smaller then or between, - toggle <'G'>, <'S'> or <'B'> respectively. The program will prompt to enter the values of one or two (if the option Between is used) thresholds. To choose values of thresholds consult the SeqWord Browser program (http://www.bi.up.ac.za/SeqWord/mhhapplet.php)
Addition of a new task
To add a new task toggle <'A'>. The program will show a new menu:
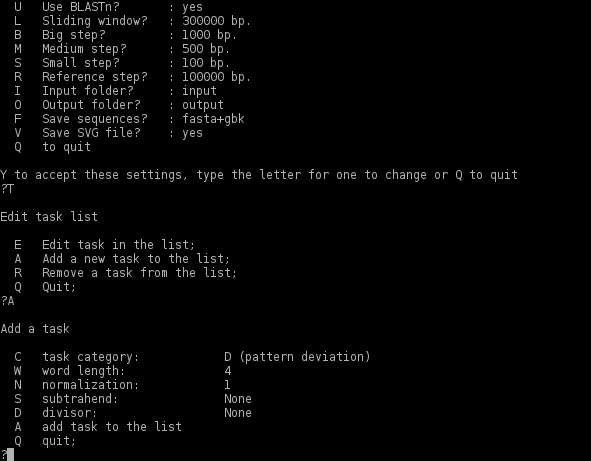
1. To choose the task category toggle <'C'> and choose from the list:
- 0. return back to the previous menu;
- 1. GRV (generalized relative variance);
- 2. PS (pattern skew);
- 3. RV (relative variance);
- 4. D (pattern deviation - by default);
- 5. GCS (GC-skew);
- 6. GD (generalized pattern deviation);
- 7. GC (GC-content);
- 8. AT (AT-content);
- 9. GPS (generalized pattern skew);
- 10. ATS (AT-skew); (for more about OU statistical parameters see Reva and Tümmler, 2005)
2. To change the oligonucleotide word length toggle <'W'> and enter an integer from 2 to 7 (4 by default).
3. To set the normalization toggle <'N'> and enter an integer from 0 (no normalization) to word_length - 1. (Normalization by the mononucleotide content of the sequence, - option 1, - is set by default. Remember, that when generalized parameters are selected, - GRV, GD or GPS, - for normalization the frequencies of the complete genome are taken into consideration, whereas by default the parameters are normalized by the content of the genomic region selected by a sliding window.)
4. The program allows execution of simple mathematical operations with the OU statistical parameters such as subtraction and division (or [par1-par2]/par3 if the subtrahend (par2) and the divisor (par3) are both set). Thus, in the scenario of identification of horizontally transferred gene islands the program calculates deviation n1_4mer:GRV/n1_4mer:RV - this ratio is around 1.0 for the core sequence but higher than 2 in genomic fragments from the accessory genome. (When setting the divisor be sure that this parameter is never zero!) To set subtraction or division of the parameters, toggle correspondingly <'S'> or <'D'>. The program will show a menu similar to the discussed above menu for addition of a new task.
Toggle <'A'> to add a subtrahend or a divisor, or to add the new task to the list. In the letter case the program will show the condition setting menu that was described above. Toggle <'Q'> to return to the task edit menu and again <'Q'> to return to the main menu.
Save a new scenario
If the list of tasks is changed, the program changes the name of the current scenario to "User defined". To save the new list of tasks in the main menu toggle <'A'> and name your scenario.
Setting the size of your sliding window
The program identifies gene islands by using a sliding window approach. To achieve optimal speed and accuracy of identification of gene islands the program flexibly changes the step of the sliding window choosing between big, medium and small steps (see below):
To change the values of the sliding window length (300 000 bp), big step (1 000 bp), medium step (500 bp) and small step (100 bp) set by default, toggle <'L'>, <'B'>, <'M'> and <'S'> respectively. The program will prompt you to enter new values. (Remember that for statistical reliability the sliding window size should not be shorter than 4600 bp for tetranucleotide usage analysis, 1200 bp for trinucleotides and 600 bp for dinucleotides.
Input and output folders
By default the program reads sequence files from the folder input and saves the result files (see an example above) to the folder output. A user may change names of the input and output folders from the main menu by toggling <'I'> and <'O'>. In addition to the text files with coordinates of identified gene islands it is possible to instruct the program to save the sequences of the gene islands to FASTA files. To do this toggle <'F'>.